Blog
Biotechnology Principles and Processes Class 12 NEET Notes

BIOTECHNOLOGY
- Biotechnology is combined term of Biology and Technology
- Biotechnology can be defined as the use of microorganisms, plants or animal cells or their components to produce products and processes useful to humans.
- Another simple definition of biotechnology is the scientific art of using living organisms to make desired products with help of technology
- According to European Federation Technology (EFT) biotechnology is the integration of natural science and organisms, cell, parts thereof and Molecular analogues for products and services.
- The term ‘Biotechnology’ was coined by Karl Ereky in 1919.
DEVELOPMENT OF BIOTECHNOLOGY
Biotechnology has developed by leaps and bounds during the past century. The development of the biotechnology can be well understood under two main heads
- Traditional biotechnology
- This technology uses bacteria and other microbes in the daily usage for preparation of dairy products. E.g. Curd, Ghee & Cheese
- This biotechnology also extends to preparation of alcoholic beverages like beer, wine etc.
- Modern biotechnology
- New biotechnology is based on rDNA technology
- Manipulates genetic information in organism. E.g. human gene producing insulin has been transferred and expressed in bacteria
PRINCIPLES OF BIOTECHNOLOGY
Genetic Engineering: Direct manipulation of genome (DNA and RNA) of an organism. It involves the transfer of new genes to improve the function or trait into host organisms and thus changes the phenotype of the host organism.
Chemical engineering: Maintenance of sterile condition to enable growth of only desired microbes for manufacture of biotechnological products like antibiotics, vaccine, enzymes etc.
TOOLS FOR GENETIC ENGINEERING
Three Types of biological tools are used in the synthesis of Recombinant DNA;
- Enzymes 2. Cloning vectors 3. Competent host
Many types of enzymes are used in genetic engineering but the most important enzymes required for genetic engineering are the lysing enzymes, restriction enzymes, DNA ligase and alkaline phosphatase
1. Enzymes
- LYSING ENZYMES
- Plant Cells: Cellulase, Pectinase, Protease & Lipase
- Animal Cells: Protease & Lipase
- Fungal Cells: Chitinase, Protease & Lipase
- Bacterial Cells: Lysozyme
- RESTRICTION ENZYMES (MOLECULAR SCISSORS)
- In 1963, Scientist discovered two enzymes responsible for restricting the growth of bacteriophage in Escherichia coli
- Enzyme 1 – It’s added methyl group to the DNA
- Enzyme 2 – It’s cut DNA at specific sites. This was later on termed as restriction endonuclease
- Restriction enzymes belong to a larger class of enzymes called Nucleases.
- There are two types of nucleases enzymes
- Exonucleases: Cut of DNA nucleotides from ends. E.g. Bal 31
- Endonucleases: Cut DNA at any point except the ends. E.g. Hind II
- Restriction enzymes are members of endonucleases
RESTRICTION ENDONUCLEASES
- First restriction endonuclease to be discovered was HindII
- The restriction enzymes always cut DNA molecules at a point of recognising a specific sequence of base pairs. This sequence is known s recognition sequence.
- Today more than 900 restriction enzymes that have been isolated from over 230 strains of bacteria with different recognition sequences.
NAMING OF RESTRICTION ENZYMES
- First letter comes from the Genus
- Second & Third letters comes from the species
- Fourth letter comes from bacterial strain
- Roman numbers after name show the order in which the enzymes were isolated from the bacterial strain.
- Examples
- Eco RI comes from Escherichia coli RY13
- Eco RII comes from Escherichia coli R 245
MECHANISM OF RESTRICTION ENZYMES
- Each restriction endonuclease recognises a specific palindromic nucleotide sequences in the DNA
- Palindrome in DNA is a. sequence of base pairs that reads same on the two strands when orientation of reading is kept same.
PLANE OF CUTTING
Blunt End
- Cut both strands of DNA through the centre
- These are known as symmetric cuts.
- As a result, two blunt ends are formed
Sticky End
- Cut in a way producing protruding and recessed ends
- These are known as asymmetric cuts.
- As a result, DNA fragments with single strand extensions formed
- ALKALINE PHOSPHATASE
- Prevention of self ligation of plasmid
- Removes phosphate groups from the 5′ end of DNA
- PHOSPHORYLASE ENZYME
- Help to again add phosphate group in DNA strands
- DNA LIGASE
- Molecular glue
- Help to formation of phosphodiester bound between 5’phosphate of one strand and 3’hydroxyl of another
- Isolated from T4 phage
2. VECTORS
- Vectors are the DNA molecules that can carry a foreign DNA segment into the host cell
- Vectors are also known as vehicle DNA or Gene carriers.
- Vectors are of two types
- Cloning Vector: used for the cloning of DNA insert inside the suitable host cell
- Expression Vector: used to express the DNA insert for producing specific protein inside the host
PROPERTIES OF VECTORS
i) Origin of replication (Ori)
- Origin of replication is a specific sequence of DNA which is responsible for initiating replication
- Any piece of DNA will be able to replicate in host only when it is integrated with his sequence
ii) Selectable marker
- They help in identifying and eliminating non transformants and selectively permit in the growth of the transformants
- Transformants: Host cells which have got Recombinant DNA
- Non transformants: Host cells which have not got Recombinant DNA
- Two types of selectable markers used in rDNA technology
- Antibiotic resistance gene E.g. Ampicillin, chloramphenicol & tetracycline
- Enzyme producing gene E.g. Beta Galoctosidase
iii) Recognition sites
- Vector should have very few recognition sites for the commonly used restriction enzymes
- Only one restriction site is preferable for an enzyme
- The presence of more than one recognition sites within the vector will generate several fragments leading to complication in gene cloning
MAJOR VECTORS USED IN rDNA TECHNOLOGY ARE:
I) PLASMIDS:
- Plasmids are small, circular, double stranded and extrachromosomal DNA present in bacterial cell
- They can replicate independently due to the presence of an origin of replication
- Plasmids are 1kbp-200kbp in size and have limited number of genes
- Plasmid vectors created from wild plasmid are called constructed vector or artificial plasmid vector
- During construction some unwanted portion removed and desired sequence are inserted
- g. pBR322,pUB & Ti Plasmid
pBR322
- This is the first artificial cloning vector
- pBR322, where p indicates that is a plasmid, B and R stands for Boliver and Rodriquez. 322 is specific number to distinguish from others
- It consist of 8 restriction sites
- pBR322 has two selectable marker genes i) Tetr gene ii) ampr gene
pUC Vectors
- This plasmid vector created by University of California are termed pUC vectors
- pUC vectors consist of an Ori Site & Lac Z gene as a selectable marker.
- Structure of all pUC vectors is same, but they differ from each other in their multiple cloning sites
Ti Plasmid
- Ti plasmids are tumor inducing plasmids present in Agrobacterium tumefaciens bacteria
- The plasmid carries transfer gene which help to transfer T-DNA from one bacterium to other bacterial or plant cell
- Ti plasmids have been used for introduction of genes of desirable traits into plants
II) BACTERIOPHAGE VECTORS
- These are bacterial virus that carries a desired gene to a host cell
- Copy number is very high that’s why it is used as cloning vector
- g. M13 Phage, Lambda Phage & P1 Phage
III) COSMIDS
- Cosmids are hybrid vectors created from Plasmid + Lambda Phage
- Cosmids gets circular DNA like Plasmid and they have cos sies like Lambda Phage
- In 1978, Casmid was first constructed by Collins and Hohn
3. HOST
- The cell receiving the rDNA is called Host
- Many types of host cells are available E.g. E.coli, yeast, animal or plant cells
- coli is the most widely used organism as its genetic engineering
- DNA is a hydrophilic molecule and it can’t pass through the plasma membrane
- DNA is a hydrophilic molecule, it can’t pass through cell membranes so the bacterial cells must first be made “competent” to take up DNA
- Therefore, certain treatments are made in the host self to become competent to take up rDNA
I) Chemical Method
- Treatment of bacterial cells with specific concentration of divalent calcium. This increase the chances of rDNA entry into the bacterial cell wall through the tiny pores
- Incubation of bacterial cells with recombinant DNA on ice. There after these are placed briefly at 42’C after this again transferred on ice. This process enable the bacteria to take up rDNA
II) Microinjection Method
- In this method, the rDNA solution is directly injected into the nucleus of animal cells
- Capillary class micropipettes or micro injections help to inject the rDNA into host cells
- This is most common in case of animal cells.
III) Gene Guns Method
- This technique is also known as a biolistic technique.
- High velocity particles of Gold or Tungsten coated with rDNA are bombarded on Host Cells
- This method is mostly used in plant cells.
IV) Electroporation method
- Electroporation is a technique used to changing the permeability of cell membrane of cells to uptake rDNA in the medium
- Electroporator instrument used to applying suitable voltage to make permeability of cell
- Electroporation is helpful to transform bacteria, fungi, plant cells and animal cells
RECOMBINANT DNA TECHNOLOGY
Recombinant DNA is a hybrid DNA formed by joining a desired foreign DNA and vector DNA
PROCESSES OF RECOMBINANT DNA TECHNOLOGY
- Isolation of DNA
- Cutting of DNA by restriction endonucleases
- Isolation of a desired Gene fragment
- Amplification of desired Gene
- Ligation of desired Gene into vector
- Transforming the recombinant DNA into the host
- Extraction of the desired product
- ISOLATION OF DNA
- First step is lysis of the cell from which the DNA has to be obtained. For this purpose various lysing enzymes are being used
- DNA is released along with other molecules like RNA, Proteins, Polysaccharides and lipids
- DNA is separated by removing other molecules from it by treating them with specific enzymes like Ribonuclease (Remove RNA) and Proteases (Remove Proteins)
- Finally, the pure DNA molecules are precipitate it out of by adding chilled ethanol and collected in the suspension.
- CUTTING OF DNA BY RESTRICTION ENDONUCLEASES
- The purified DNA is incubated with the specific restriction enzyme at conditions optimum for the enzyme to act.
- Also, a vector DNA treated with the same specific restriction enzyme
- Example: If cut purified DNA with EcoR1 and same enzyme used to cut vector DNA also
- ISOLATION OF A DESIRED GENE
- Desired DNA fragments is separated by using agarose gel electrophoresis technique
GEL ELECTROPHORESIS
- A restriction endonucleases enzyme cuts DNA into small fragments.
- These fragments are separated by gel electrophoresis
- Gel electrophoresis is a technique, which separates DNA fragments based on their size
- DNA fragments are negatively charged molecules. They can be separated by forcing them to move towards the anode under an electric field through a medium
- The most common medium used is agarose, a natural polymer extracted from sea weeds
- The smaller fragments move farther away as compared to larger fragments.
- The DNA fragments are strained with its Ethridium Bromide for visualization.
- The bright orange colour DNA bands can be seen after the exposure of UV light.
- These bands then are cut out from the gel and extracted by using a convenient technique. This step is called Elution.
- DNA fragments are then purified and then used for construction of Recombinant DNA
- AMPLIFICATION OF DESIRED GENE
- Polymerase chain reaction is a technique of synthesizing multiple copies of the desired genes a DNA.
- It was developed by Karry Mullis.
POLYMERASE CHAIN REACTION
REQUIREMENTS OF PCR:
- DNA Template: The desired segment of the target DNA that is to be amplified.
- Primers: These are oligonucleotides of 10 to 8 nucleotides that are complementary to DNA template region.
- Enzyme: High temperature (>90°C) stable DNA polymerase (usually Taq Polymerase) is used for synthesis of new DNA molecule required. Taq polymerase is obtained from Thermusaquaticus bacterium.
MECHANISM OF PCR:
- Denaturation: Target DNA is heated to high temperature of about 95°C for 1 minute. This results in the separation of the two strands of the target double stranded DNA.
- Annealing: It is carried out by Primers, which are added in the reaction. Primers aneal or join to the three prime end of each separated strands. It occurs at 55°C or 30 seconds.
- Extension: It is done by DNA polymerase by adding nucleotides complementary to the template in the reaction. It occurs at 72°C for 1 minute.
- These steps are repeated for many times to obtain several copies of the desired DNA
- LIGATION OF DESIRED GENE INTO VECTOR
- The gene of interest or desired Gene is ligated with the vector DNA with the help of enzyme DNA ligase to form Recombinant DNA.
- TRANSFORMING THE RECOMBINANT DNA INTO THE HOST
- Recipient cells are made competent by some technique which has already been discussed previously
- For example, if a Recombinant DNA bearing antibiotic ampicillin resistant gene is inserted in E.coli cells the bacteria becomes resistant to ampicillin. If such bacteria are transferred on culture plate containing ampicillin, only resistant forms bacteria will grow. And others bacteria will die. This method is used to separate transformants and non transformants. The gene responsible for ampicillin resistance is known as selectable marker
- EXTRACTION OF THE DESIRED PRODUCT
- The foreign DNA multiplies in Host cell to produce desirable protein.
- This expression of the foreign gene requires appropriate conditions in host cell
- Coding genes expressed in a heterologous host is called Recombinant protein.
- Small scale production can be done through culturing in laboratory
- For producing very high yields of the desired proteins, Bioreactors are required
BIOREACTORS
- Bioreactors are an instrument. These are large volume (100-1000L) vessels in which raw materials are biologically converted into specific products, individual enzymes, etc. using microbial plants animals or human cells.
- It provides optimum conditions for achieving the desired product by providing optimum growth conditions like temperature, pH, salt, substrate, vitamins and oxygen.
- Bioreactors are two types:
- STIRRED BIOREACTOR: These are cylindrical shaped and facilities mixing of reactor contents and checks oxygen availability throughout the bioreactor.
- SPARGED STIRRED BIOREACTOR: Sterile air Bubbles are passed, to increase the surface area for oxygen transfer.
COMPONENTS OF BIOREACTOR:
- Agitator system
- Oxygen delivery system
- Foam control system
- Temperature and pH control system
- Sampling ports to extract small volumes of culture
DOWNSTREAM PROCESSING:
After the product formation in bioreactors, they are subjected to a series of processes, collectively called downstream processes. This includes
- Separation of desired products
- Verification of products
- Formulation with preservatives
These products undergo strict quality control testing before being marketed as finished products.

 Blog8 months ago
Blog8 months ago[PPT] Human Reproduction Class 12 Notes
- Blog8 months ago
Contribution of Indian Phycologists (4 Famous Algologist)
- Blog8 months ago
PG TRB Botany Study Material PDF Free Download

 Blog8 months ago
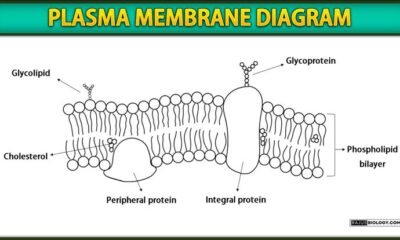
Blog8 months agoPlasma Membrane Structure and Functions | Free Biology Notes

 Blog8 months ago
Blog8 months agoCell The Unit of Life Complete Notes | Class 11 & NEET Free Notes

 Blog8 months ago
Blog8 months ago[PPT] The living world Class 11 Notes

 Blog8 months ago
Blog8 months agoClassification of Algae By Fritsch (11 Classes of Algae)

 Blog8 months ago
Blog8 months agoJulus General Characteristics | Free Biology Notes